
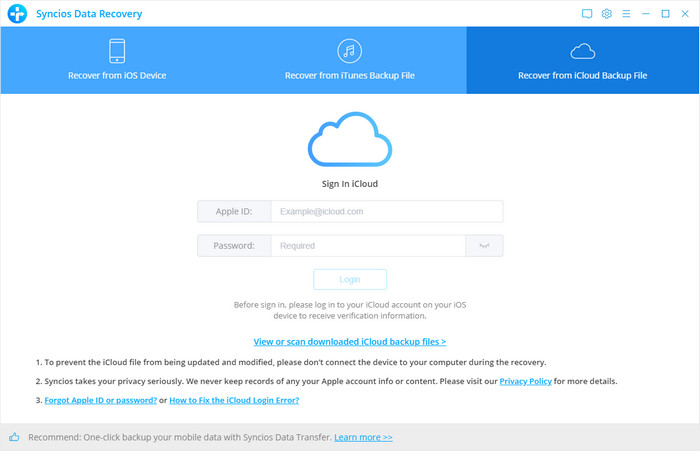
#Syncios data recovery manual
Manual gating using analyzer-defined boundaries or “gates” to identify cell populations of interest is commonly analyzed using proprietary software (e.g. We describe a practical method for standardization of immunophenotyping methods in large scale population studies that provides a rapid, accurate and reproducible alternative to labor intensive manual gating strategies.įlow cytometry (FCM) provides a high dimensional quantitative measure for single cell analysis of the immune system.
The mean difference in the absolute counts between the hybrid method and manual gating strategy of these cell subsets showed results that were very similar to the traditional manual gating method. For the remaining subsets, the correlation was low (<0.80) primarily because of the low numbers of events recorded in these subsets. Our results show a high degree of correlation (r ≥ 0.80) for 18 (78%) of the 24 cell subsets. We validated this method with traditional manual gating strategies for 24 subsets of T cells, B cells, NK cells, monocytes and dendritic cells in 931 participants from the Health and Retirement Study (HRS). This approach allows pre-defining populations that can be analyzed solely by automated analysis and incorporating manual refinement for smaller downstream populations. Therefore, we developed an approach that utilizes a previously published automated algorithm (OpenCyto framework) with a manually created hierarchically cell gating template implemented, along with a custom developed visualization software (FlowAnnotator) to rapidly and efficiently analyze immunophenotyping data in large population studies.

However, many of the automated algorithms still require significant manual interventions or have yet to demonstrate their utility in large datasets. Several automated gating methods have shown to exceed performance of manual gating for a limited number of cell subsets. Traditional manual gating strategies are often time-intensive, place a high burden on the analyzer, and are susceptible to bias between analyzers.


 0 kommentar(er)
0 kommentar(er)
